Overview of my research:
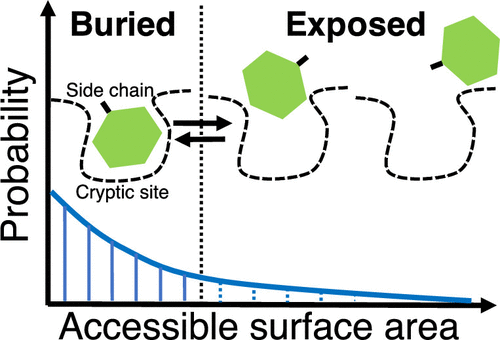
Cryptic Binding Site of Proteins
We have found unique side-chain fluctuations of cryptic binding sites via molecular dynamics simulations.
Source code to analyse the fluctuations 👉 github
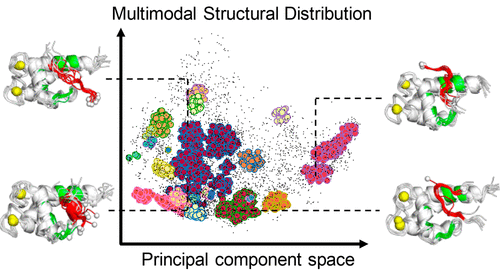
Intrinsically Disordererd Protein
We have explored the structural ensemble of an intrinsically disordered region, p53 C-terminal domain, via a generalised ensemble molecular dynamics simulation. We have identified various binding modes on a target protein.
Our papers 👇
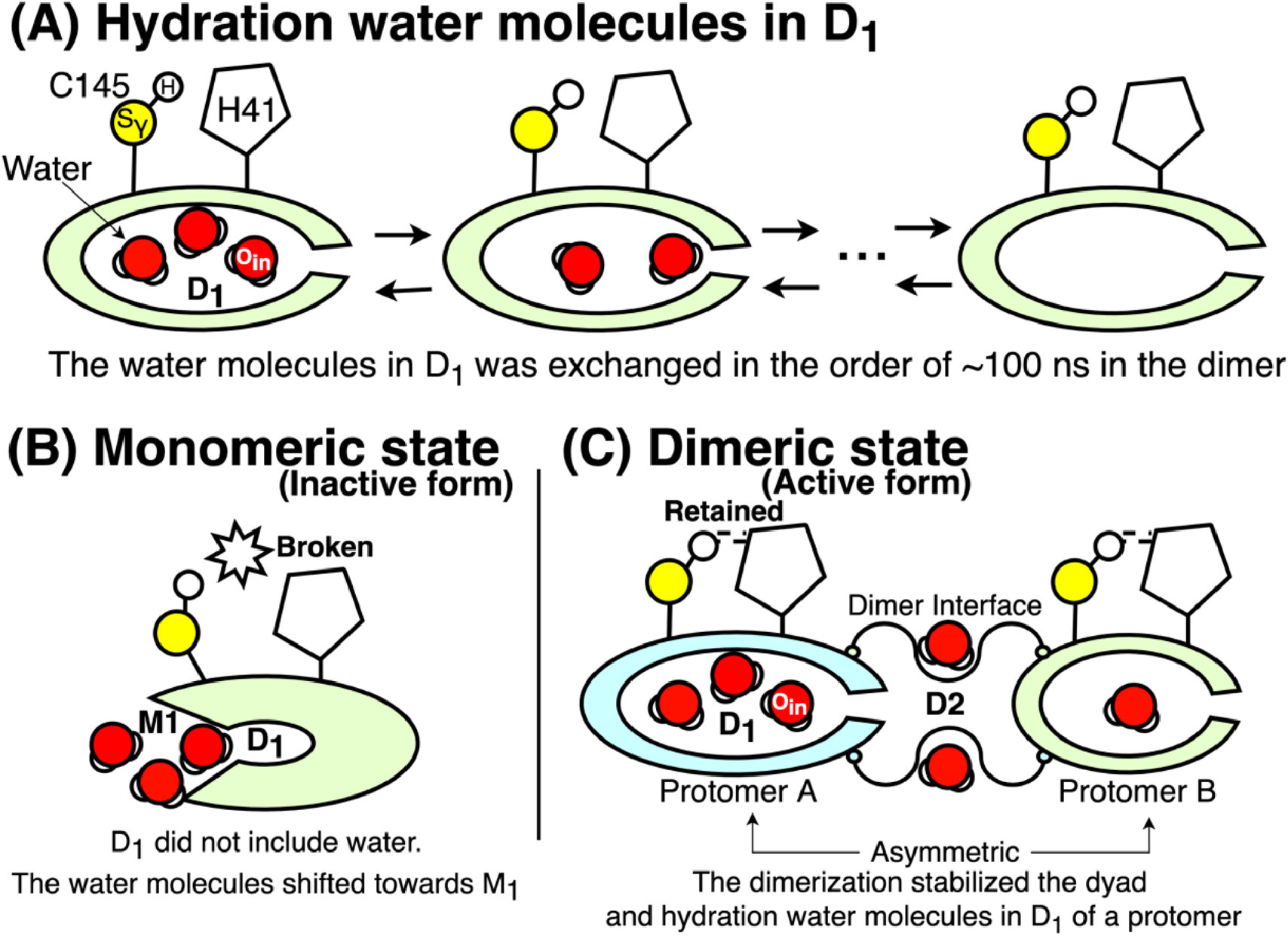
SARS-CoV-2 main protease
We have investigated monomeric and dimeric states of the SARS-CoV-2 main protease, identifying water molecules that can be crucial to keep the dyad configuration.
Written by Shinji Iida. Last modified: May 08, 2022

 Google Scholar
Google Scholar DockerHub
DockerHub